tune_grid() computes a set of performance metrics (e.g. accuracy or RMSE)
for a pre-defined set of tuning parameters that correspond to a model or
recipe across one or more resamples of the data.
Usage
tune_grid(object, ...)
# S3 method for class 'model_spec'
tune_grid(
object,
preprocessor,
resamples,
...,
param_info = NULL,
grid = 10,
metrics = NULL,
eval_time = NULL,
control = control_grid()
)
# S3 method for class 'workflow'
tune_grid(
object,
resamples,
...,
param_info = NULL,
grid = 10,
metrics = NULL,
eval_time = NULL,
control = control_grid()
)Arguments
- object
A
parsnipmodel specification or an unfitted workflow(). No tuning parameters are allowed; if arguments have been marked with tune(), their values must be finalized.- ...
Not currently used.
- preprocessor
A traditional model formula or a recipe created using
recipes::recipe().- resamples
An
rsetresampling object created from anrsamplefunction, such asrsample::vfold_cv().- param_info
A
dials::parameters()object orNULL. If none is given, a parameters set is derived from other arguments. Passing this argument can be useful when parameter ranges need to be customized.- grid
A data frame of tuning combinations or a positive integer. The data frame should have columns for each parameter being tuned and rows for tuning parameter candidates. An integer denotes the number of candidate parameter sets to be created automatically.
- metrics
A
yardstick::metric_set(), orNULLto compute a standard set of metrics.- eval_time
A numeric vector of time points where dynamic event time metrics should be computed (e.g. the time-dependent ROC curve, etc). The values must be non-negative and should probably be no greater than the largest event time in the training set (See Details below).
- control
An object used to modify the tuning process, likely created by
control_grid().
Value
An updated version of resamples with extra list columns for .metrics and
.notes (optional columns are .predictions and .extracts). .notes
contains warnings and errors that occur during execution.
Details
Suppose there are m tuning parameter combinations. tune_grid() may not
require all m model/recipe fits across each resample. For example:
In cases where a single model fit can be used to make predictions for different parameter values in the grid, only one fit is used. For example, for some boosted trees, if 100 iterations of boosting are requested, the model object for 100 iterations can be used to make predictions on iterations less than 100 (if all other parameters are equal).
When the model is being tuned in conjunction with pre-processing and/or post-processing parameters, the minimum number of fits are used. For example, if the number of PCA components in a recipe step are being tuned over three values (along with model tuning parameters), only three recipes are trained. The alternative would be to re-train the same recipe multiple times for each model tuning parameter.
tune supports parallel processing with the future package. To execute
the resampling iterations in parallel, specify a plan with
future first. The allow_par argument can be used to avoid parallelism.
For the most part, warnings generated during training are shown as they occur
and are associated with a specific resample when
control_grid(verbose = TRUE). They are (usually) not aggregated until the
end of processing.
Parameter Grids
If no tuning grid is provided, a grid (via dials::grid_space_filling()) is
created with 10 candidate parameter combinations.
When provided, the grid should have column names for each parameter and
these should be named by the parameter name or id. For example, if a
parameter is marked for optimization using penalty = tune(), there should
be a column named penalty. If the optional identifier is used, such as
penalty = tune(id = 'lambda'), then the corresponding column name should
be lambda.
In some cases, the tuning parameter values depend on the dimensions of the
data. For example, mtry in random forest models depends on the number of
predictors. In this case, the default tuning parameter object requires an
upper range. dials::finalize() can be used to derive the data-dependent
parameters. Otherwise, a parameter set can be created (via
dials::parameters()) and the dials update() function can be used to
change the values. This updated parameter set can be passed to the function
via the param_info argument.
The rows of the grid are called tuning parameter candidates. Each
candidate has a unique .config value that, for grid search, has the
pattern pre{num}_mod{num}_post{num}. The numbers include a zero when that
element was static. For example, a value of pre0_mod3_post4 means no
preprocessors were tuned and the model and postprocessor(s) had at least
three and four candidates, respectively. Also, the numbers are zero-padded
to enable proper sorting.
Performance Metrics
To use your own performance metrics, the yardstick::metric_set() function
can be used to pick what should be measured for each model. If multiple
metrics are desired, they can be bundled. For example, to estimate the area
under the ROC curve as well as the sensitivity and specificity (under the
typical probability cutoff of 0.50), the metrics argument could be given:
metrics = metric_set(roc_auc, sens, spec)Each metric is calculated for each candidate model.
If no metric set is provided, one is created:
For regression models, the root mean squared error and coefficient of determination are computed.
For classification, the area under the ROC curve and overall accuracy are computed.
Note that the metrics also determine what type of predictions are estimated during tuning. For example, in a classification problem, if metrics are used that are all associated with hard class predictions, the classification probabilities are not created.
The out-of-sample estimates of these metrics are contained in a list column
called .metrics. This tibble contains a row for each metric and columns
for the value, the estimator type, and so on.
collect_metrics() can be used for these objects to collapse the results
over the resampled (to obtain the final resampling estimates per tuning
parameter combination).
Obtaining Predictions
When control_grid(save_pred = TRUE), the output tibble contains a list
column called .predictions that has the out-of-sample predictions for each
parameter combination in the grid and each fold (which can be very large).
The elements of the tibble are tibbles with columns for the tuning
parameters, the row number from the original data object (.row), the
outcome data (with the same name(s) of the original data), and any columns
created by the predictions. For example, for simple regression problems, this
function generates a column called .pred and so on. As noted above, the
prediction columns that are returned are determined by the type of metric(s)
requested.
This list column can be unnested using tidyr::unnest() or using the
convenience function collect_predictions().
Extracting Information
The extract control option will result in an additional function to be
returned called .extracts. This is a list column that has tibbles
containing the results of the user's function for each tuning parameter
combination. This can enable returning each model and/or recipe object that
is created during resampling. Note that this could result in a large return
object, depending on what is returned.
The control function contains an option (extract) that can be used to
retain any model or recipe that was created within the resamples. This
argument should be a function with a single argument. The value of the
argument that is given to the function in each resample is a workflow
object (see workflows::workflow() for more information). Several
helper functions can be used to easily pull out the preprocessing
and/or model information from the workflow, such as
extract_preprocessor() and
extract_fit_parsnip().
As an example, if there is interest in getting each parsnip model fit back, one could use:
extract = function (x) extract_fit_parsnip(x)Note that the function given to the extract argument is evaluated on
every model that is fit (as opposed to every model that is evaluated).
As noted above, in some cases, model predictions can be derived for
sub-models so that, in these cases, not every row in the tuning parameter
grid has a separate R object associated with it.
Finally, it is a good idea to include calls to require() for packages that
are used in the function. This helps prevent failures when using parallel
processing.
Case Weights
Some models can utilize case weights during training. tidymodels currently supports two types of case weights: importance weights (doubles) and frequency weights (integers). Frequency weights are used during model fitting and evaluation, whereas importance weights are only used during fitting.
To know if your model is capable of using case weights, create a model spec
and test it using parsnip::case_weights_allowed().
To use them, you will need a numeric column in your data set that has been
passed through either hardhat:: importance_weights() or
hardhat::frequency_weights().
For functions such as fit_resamples() and the tune_*() functions, the
model must be contained inside of a workflows::workflow(). To declare that
case weights are used, invoke workflows::add_case_weights() with the
corresponding (unquoted) column name.
From there, the packages will appropriately handle the weights during model fitting and (if appropriate) performance estimation.
Censored Regression Models
Three types of metrics can be used to assess the quality of censored regression models:
static: the prediction is independent of time.
dynamic: the prediction is a time-specific probability (e.g., survival probability) and is measured at one or more particular times.
integrated: same as the dynamic metric but returns the integral of the different metrics from each time point.
Which metrics are chosen by the user affects how many evaluation times should be specified. For example:
# Needs no `eval_time` value
metric_set(concordance_survival)
# Needs at least one `eval_time`
metric_set(brier_survival)
metric_set(brier_survival, concordance_survival)
# Needs at least two eval_time` values
metric_set(brier_survival_integrated, concordance_survival)
metric_set(brier_survival_integrated, concordance_survival)
metric_set(brier_survival_integrated, concordance_survival, brier_survival)Values of eval_time should be less than the largest observed event
time in the training data. For many non-parametric models, the results beyond
the largest time corresponding to an event are constant (or NA).
Examples
library(recipes)
library(rsample)
library(parsnip)
library(workflows)
library(ggplot2)
# ---------------------------------------------------------------------------
set.seed(6735)
folds <- vfold_cv(mtcars, v = 5)
# ---------------------------------------------------------------------------
# tuning recipe parameters:
spline_rec <-
recipe(mpg ~ ., data = mtcars) |>
step_spline_natural(disp, deg_free = tune("disp")) |>
step_spline_natural(wt, deg_free = tune("wt"))
lin_mod <-
linear_reg() |>
set_engine("lm")
# manually create a grid
spline_grid <- expand.grid(disp = 2:5, wt = 2:5)
# Warnings will occur from making spline terms on the holdout data that are
# extrapolations.
spline_res <-
tune_grid(lin_mod, spline_rec, resamples = folds, grid = spline_grid)
spline_res
#> # Tuning results
#> # 5-fold cross-validation
#> # A tibble: 5 × 4
#> splits id .metrics .notes
#> <list> <chr> <list> <list>
#> 1 <split [25/7]> Fold1 <tibble [32 × 6]> <tibble [0 × 4]>
#> 2 <split [25/7]> Fold2 <tibble [32 × 6]> <tibble [0 × 4]>
#> 3 <split [26/6]> Fold3 <tibble [32 × 6]> <tibble [0 × 4]>
#> 4 <split [26/6]> Fold4 <tibble [32 × 6]> <tibble [0 × 4]>
#> 5 <split [26/6]> Fold5 <tibble [32 × 6]> <tibble [0 × 4]>
show_best(spline_res, metric = "rmse")
#> # A tibble: 5 × 8
#> disp wt .metric .estimator mean n std_err .config
#> <int> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
#> 1 3 2 rmse standard 2.54 5 0.207 pre05_mod0_post0
#> 2 3 3 rmse standard 2.64 5 0.234 pre06_mod0_post0
#> 3 4 3 rmse standard 2.82 5 0.456 pre10_mod0_post0
#> 4 4 2 rmse standard 2.93 5 0.489 pre09_mod0_post0
#> 5 4 4 rmse standard 3.01 5 0.475 pre11_mod0_post0
# ---------------------------------------------------------------------------
# tune model parameters only (example requires the `kernlab` package)
car_rec <-
recipe(mpg ~ ., data = mtcars) |>
step_normalize(all_predictors())
svm_mod <-
svm_rbf(cost = tune(), rbf_sigma = tune()) |>
set_engine("kernlab") |>
set_mode("regression")
# Use a space-filling design with 7 points
set.seed(3254)
svm_res <- tune_grid(svm_mod, car_rec, resamples = folds, grid = 7)
svm_res
#> # Tuning results
#> # 5-fold cross-validation
#> # A tibble: 5 × 4
#> splits id .metrics .notes
#> <list> <chr> <list> <list>
#> 1 <split [25/7]> Fold1 <tibble [14 × 6]> <tibble [0 × 4]>
#> 2 <split [25/7]> Fold2 <tibble [14 × 6]> <tibble [0 × 4]>
#> 3 <split [26/6]> Fold3 <tibble [14 × 6]> <tibble [0 × 4]>
#> 4 <split [26/6]> Fold4 <tibble [14 × 6]> <tibble [0 × 4]>
#> 5 <split [26/6]> Fold5 <tibble [14 × 6]> <tibble [0 × 4]>
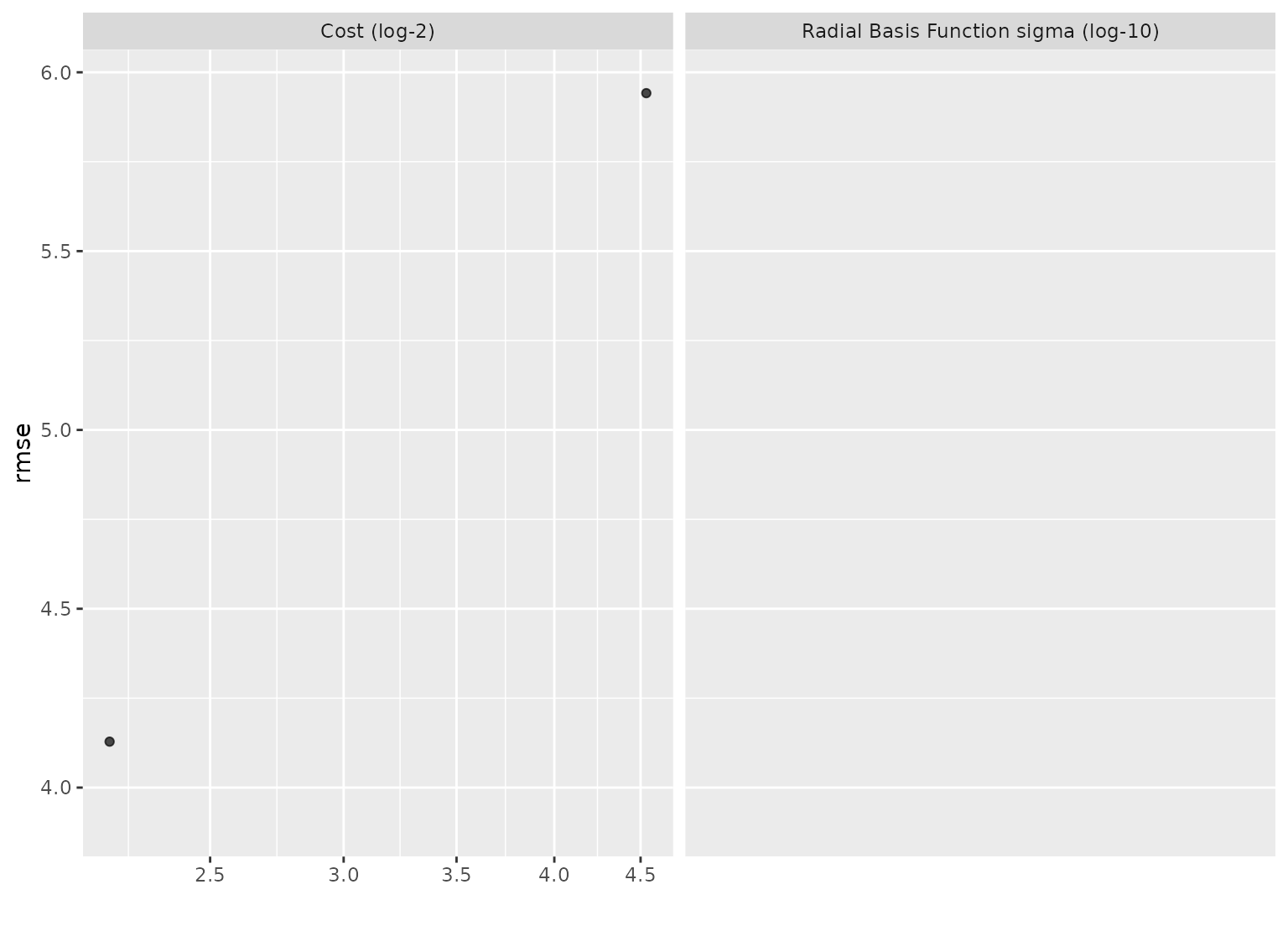
show_best(svm_res, metric = "rmse")
#> # A tibble: 5 × 8
#> cost rbf_sigma .metric .estimator mean n std_err .config
#> <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
#> 1 5.66 0.0215 rmse standard 2.69 5 0.186 pre0_mo…
#> 2 0.0312 1 rmse standard 5.82 5 0.946 pre0_mo…
#> 3 32 0.000000215 rmse standard 5.95 5 0.969 pre0_mo…
#> 4 0.177 0.00001 rmse standard 5.96 5 0.970 pre0_mo…
#> 5 0.000977 0.000464 rmse standard 5.96 5 0.970 pre0_mo…
autoplot(svm_res, metric = "rmse") +
scale_x_log10()
#> Warning: NaNs produced
#> Warning: log-10 transformation introduced infinite values.
#> Warning: Removed 10 rows containing missing values or values outside the scale
#> range (`geom_point()`).
 # ---------------------------------------------------------------------------
# Using a variables preprocessor with a workflow
# Rather than supplying a preprocessor (like a recipe) and a model directly
# to `tune_grid()`, you can also wrap them up in a workflow and pass
# that along instead (note that this doesn't do any preprocessing to
# the variables, it passes them along as-is).
wf <- workflow() |>
add_variables(outcomes = mpg, predictors = everything()) |>
add_model(svm_mod)
set.seed(3254)
svm_res_wf <- tune_grid(wf, resamples = folds, grid = 7)
# ---------------------------------------------------------------------------
# Using a variables preprocessor with a workflow
# Rather than supplying a preprocessor (like a recipe) and a model directly
# to `tune_grid()`, you can also wrap them up in a workflow and pass
# that along instead (note that this doesn't do any preprocessing to
# the variables, it passes them along as-is).
wf <- workflow() |>
add_variables(outcomes = mpg, predictors = everything()) |>
add_model(svm_mod)
set.seed(3254)
svm_res_wf <- tune_grid(wf, resamples = folds, grid = 7)
